Group | Genotyping
-
Medicine
-
Medicines Development Unit
- Diagnostic Development
- Genotyping
- Molecularimmunology
-
Medicines Development Unit
Malaria molecular surveillance (MMS) is increasingly being implemented in malaria-endemic countries and provides useful information to complement traditional epidemiological data. Indeed, the use of adequate and highly sensitive techniques for PCR correction, monitoring of drug and insecticide resistance markers or histidine-rich protein 2 and 3 deletions are key to effective malaria interventions. In low transmission settings, the detection of asymptomatic carriers and characterisation of transmission chains are crucial for reactive case interventions aiming at malaria elimination.
Research
The Genotyping Group develops and implements molecular assays for molecular surveillance of malaria. This include Next Generation Sequencing (NGS) assays for targeted amplicon deep sequencing (TADS) and whole genome sequencing (WGS). Current projects include the investigation into alternative genotyping methods, including microsatellites and amplicon sequencing, to improve our understanding of parasite population dynamics and current methods for distinguishing recrudescence from new infection, and the identification of new molecular markers of drug resistance. We are working closely with National Malaria Control Programmes (NMCPs) and research institutes in malaria-endemic countries to implement these new assays into routine surveillance and provide critical evidence for malaria monitoring, particularly for mitigating antimalarial drug resistance. Together with other groups at Swiss TPH, we support elimination efforts by using genomic and serological techniques to understand parasite reservoirs and guide the implementation of targeted elimination interventions.
Services provision
Genotyping services are provided to drug developers performing regulatory clinical trials. Our laboratory is accredited to ISO 17025:2017 and is increasingly implementing a GxP compliant quality management system. Services provided include:
- Distinguishing recrudescence from new infection,
- Identification of antimalarial drug resistance markers,
- Quantitative PCRs for low parasite density, and
- Detection and quantification of gametocytes

Christian Nsanzabana
PD, PhD, Dr.
Group Leader
+41612848252
,
+41612848237
christian.nsanzabana@swisstph.ch
Key Projects

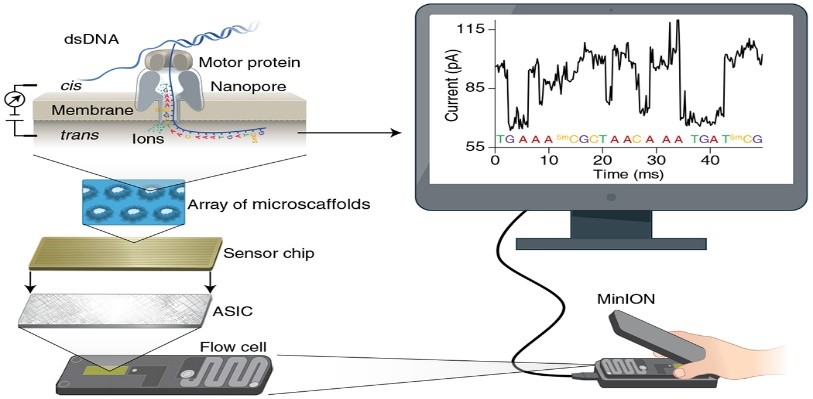

SPREAD - Assessment of the Factors Affecting Emergence and Spread of Artemisinin Resistance in Rwanda
Artemisinin resistance has been reported and confirmed in Rwanda and other countries in East and Horn of Africa; calling for a thorough investigation to understand the factors affecting the emergence of resistance and spread in the African context. In this project, NMCP staff will be trained on molecular techniques and ex vivo assays, to better understand the factors affecting the emergence and spread of artemisinin resistance in Rwanda. This project is aiming at strengthening the capacity for antimalarial drug resistance surveillance in Rwanda, providing robust data to the national health authorities to monitor the spread of artemisinin and partner dug resistance. Read more
Genome-wide Analysis of Variations in Plasmodium falciparum Parasite Populations from Different Regions of Tanzania
The objective of the project is to develop a whole genome sequencing (WGS) assay based on long-reads with Oxford Nanopore technology (ONT - Minion), through exploring different DNA extraction and DNA enrichment methods prior to ONT sequencing. The study is aiming at identifying the ideal laboratory assay for sufficient DNA yield required for WGS, and optimizing the recommended method for generation of long-reads data. This project is part of a large consortium; the Molecular Surveillance of Malaria in Tanzania (MSMT) Project led by the National Institute of Medical Research (NIMR) in Tanzania. The assay developed will be transferred to NIMR laboratory in Tanzania. Through this study, new variants may be discovered, which could be used in surveillance and monitoring the emergence and spread of drug resistance and hrp2/3 gene deletions. Read more
Evaluation of the Community-based Malaria Surveillance and Response in Rwanda
Malaria incidence has considerably reduced in Rwanda in the last 5 years through combined malaria control interventions. Further decreasing and reducing transmissions to zero in our communities including high-risk or vulnerable populations is essential to achieving and sustaining malaria elimination. To achieve this, the research project proposes to (1) evaluate the impact of malaria control interventions in Rwanda over the last 5 years, (2) evaluate the contribution of integrated interventions approach through reactive Community-based Malaria Surveillance and Response in accelerating malaria elimination in Rwanda and (3) identify malaria infection characteristics and risk factors in Rwanda to inform tailored interventions. Read more
Latest Publications
All PublicationsGolumbeanu M, Edi C.A.V, Hetzel M.W, Koepfli C, Nsanzabana C. Bridging the gap from molecular surveillance to programmatic decisions for malaria control and elimination. Am J Trop Med Hyg. 2025;112(1 Suppl.):35-47. DOI: 10.4269/ajtmh.22-0749
Kweyamba P.A et al. Contrasting vector competence of three main East African Anopheles malaria vector mosquitoes for Plasmodium falciparum. Sci Rep. 2025;15:2286. DOI: 10.1038/s41598-025-86409-w
Mwesigwa A et al. Assessment of different genotyping markers and algorithms for distinguishing Plasmodium falciparum recrudescence from reinfection in Uganda. Sci Rep. 2025;15:4375. DOI: 10.1038/s41598-025-88892-7
Bakari C et al. Trends of Plasmodium falciparum molecular markers associated with resistance to artemisinins and reduced susceptibility to lumefantrine in Mainland Tanzania from 2016 to 2021. Malar J. 2024;23:71. DOI: 10.1186/s12936-024-04896-0
Hofer L.M et al. Additional blood meals increase sporozoite infection in Anopheles mosquitoes but not Plasmodium falciparum genetic diversity. Sci Rep. 2024;14:17467. DOI: 10.1038/s41598-024-67990-y
Holzschuh A et al. Using a mobile nanopore sequencing lab for end-to-end genomic surveillance of Plasmodium falciparum: a feasibility study. PLOS Glob Public Health. 2024;4(2):e0002743. DOI: 10.1371/journal.pgph.0002743
 Nukunu Akyea-Bobi
Nukunu Akyea-Bobi
 Anara Alshanbayeva
Anara Alshanbayeva
 Patricia Bravo
Patricia Bravo
 Sara Cantoreggi
Sara Cantoreggi
 Lorenz Hofer
Lorenz Hofer
 Aurel Holzschuh
Aurel Holzschuh
 Daniela Montero Salas
Daniela Montero Salas
 Christian Nsanzabana
Christian Nsanzabana
 Kelsey Schärer
Kelsey Schärer
 Michaela Zwyer
Michaela Zwyer